Dinoflagellate Rhodopsin: A Proton Pump to Harness Light Energy:
Animal vision relies on rhodopsin (protein) and its cofactor 11-cis retinal. It turns out many microbes in the aquatic ecosystem also possess rhodopsin. Microbial rhodopsin is diverse genetically and functionally. Although some are also light sensing (e.g. for phototaxis), most of the microbial rhodopsins produce a motive force (e.g. driving flagellum rotation) or generate ATP (Adenosine Triphosphate), the energy currency in living cells, from light. The latter type, with all-trans retinal as the cofactor, is named proton pump type rhodopsin (PPR), because it generates a proton gradient across the membrane leading to ATP production. PPR augments a photosynthetic apparatus for harvesting solar energy to produce ATP. This has significant ecological implications because phototrophy is the ultimate source of energy that drives the function of an ecosystem. PPR occurs widely in in ocean surface, it is found in 13% to 80% of marine bacteria and archaea (Béjà et al. 2001, de la Torre et al. 2003, Moran and Miler 2007). These photoproteins seems to be specifically tuned to absorb the wavelengths of light found in the surrounding environment: green light in surface waters and blue light at greater depths in the water columns.
The presence and potential functions of rhodopsin in marine eukaryotic protists (phytoplankton and heterotrophic counterparts), which are the major player in marine biogeochemical processes, is still poorly understood. Rhodopsin genes have been identified in some fungi and eukaryotic microalgae, some of these are apparently proton pumps like PR, as in Leptosphaeria (Waschuk et al. 2005), while others are sensors involved in phototaxis, as in Chlamydomonas (Foster et al. 1984, Sineshchekov et al. 2002). Recent research shows that PPR seem to be particularly widespread in different lineages of dinoflagellates: based on transcriptomic analyses on lab cultures and natural assemblages, rhodopsin has now been found in the ancestral and heterotrophic species O. marina, later-diverging peridinin-plastid species Polarella antartica, Pyrocystis lunulla, Alexandrium catenella and Karlodinium veneficum, as well as a natural dinoflagellate assemblage in Long Island Sound (Zhang et al. 2007, Lin et al. 2010).
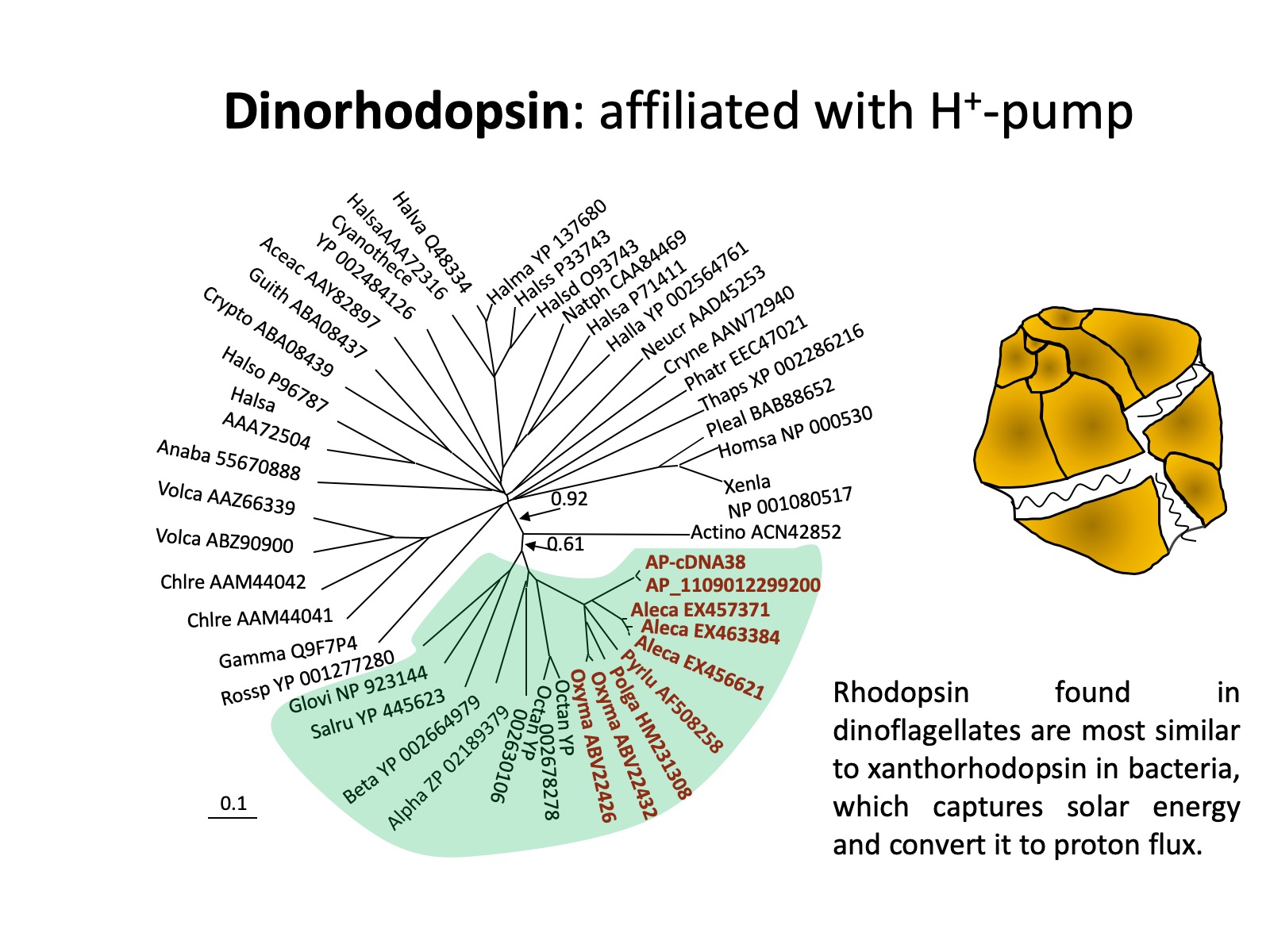
It also has been identified in cryptophytes and diatoms (Marchetti et al. 2011). Further, comparative analysis on the sequences from dinoflagellates and other organisms shows that dinoflagellate rhodopsin is most closely related to xanthorhodopsin, a subtype of PPR, with particular high affinity to subgroup II, which does not (in contrast to subgroup I that does) contain a carotenoid molecule as well as retinal in the light-harvesting antenna. The presence of rhodopsin in dinoflagellates has subsequently been confirmed with three types of rhodopsin genes found in the phylum of dinoflagellates (Slamovits et al. 2011). The dinoflagellate rhodopsin possesses all the typical transmembrane domains, the amino acid residues conserved for ion-type rhodopsin and the residue that is critical for proton pump.
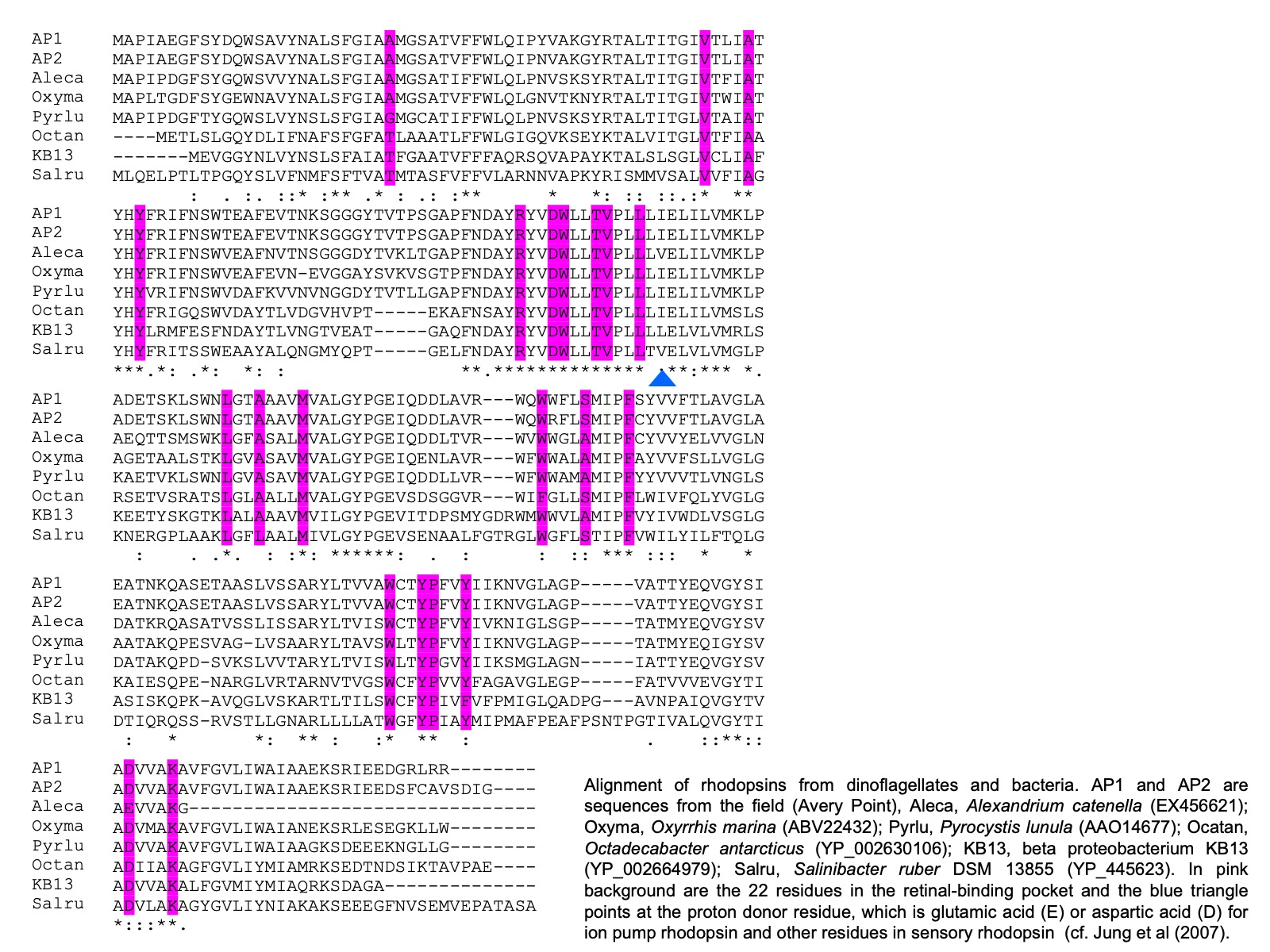
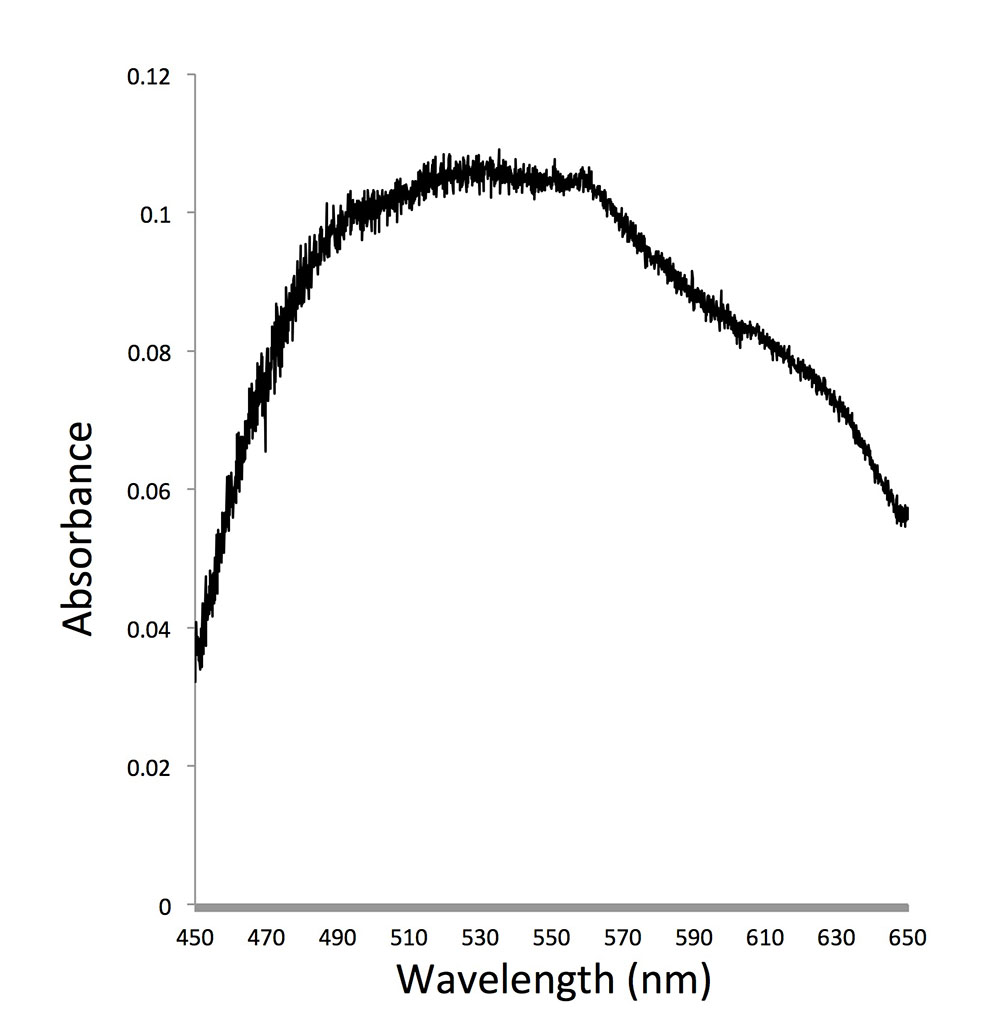
As dinoflagellates are one of the most important groups of primary producers and micrograzers in the ocean, the role of dinoflagellate rhodopsin can have profound impact on the function of the marine ecosystem. We try to understand whether this protein indeed absorbs light and converts it to ATP in both photosynthetic and heterotrophic dinoflagellates and how common (and diverse) this gene is in the dinoflagellate communities. Regarding the first question, expression of PPR in starved O. marina kept under light led to enhanced survival compared to those kept in the dark. In addition, similar to proteorhodopsin in bacteria, we found that rhodopsin in dinoflagellates had optimal absorption in the blue to green spectrum. These results suggest that dinoflagellate rhodopsins most likely function as proton pump to support growth of dinoflagellates under nutrient-limited conditions.
The second question was addressed by cloning and sequencing rhodopsin genes using primers designed from conserved sequence regions of dinoflagellate rhodopsins. We found that dinoflagellate rhodopsin like sequences were highly diverse in Long Island Sound (LIS). Total abundance of dinoflagellate rhodopsin like sequences fluctuated seasonally. These not yet published results suggest that dinoflagellate rhodopsins may play some roles in sustaining dinoflagellate populations.
Our laboratory trains students at all levels on rhodopsin study, ranging from high school to postdoctoral levels. These students, including female and minority populations, play important roles in the research leading to the understanding achieved in our laboratory so far. Besides, some of the new understanding has been incorporated into some courses taught by me. In 2011, students who took the course MARN5015: Molecular Approaches to Biological Oceanography were directly involved in the analysis of dinoflagellate rhodopsin diversity in LIS.