Dinoflagellates are major contributors of symbioses as well as harmful algal blooms (HABs) in the ocean. They form symbiotic relationships with a wide range of invertebrates (corals, giant clams, forams, jellyfish), in which species of the Symbiodiniaceae family function as symbionts, and with algae (e.g. diatoms, prasinophytes), in which species such as Kryptoperidinium and Notciluca serve as hosts. For genomic database and analysis resources for dinoflagellates and relevant algae pertaining to symbioses, please visit the SAGER website.

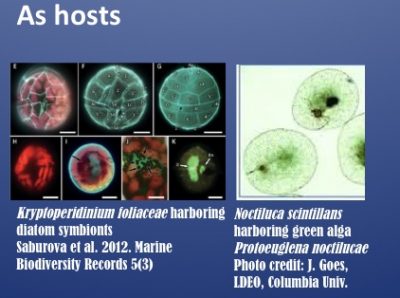
The symbiotic Noctiluca scintillans forms massive green blooms in the Arabian Sea, imposing significant ecological and economic impacts.

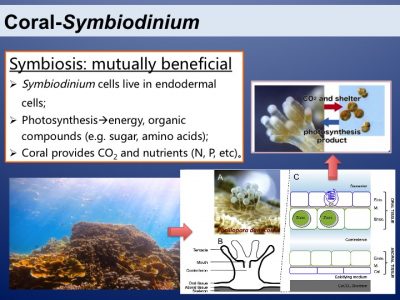
Coral reefs are the most productive and biodiverse marine ecosystems. This is because of the amazing symbiotic relationship between corals and the dinoflagellates from the family of Symbiodiniaceae. The two symbiotic partners benefit mutually. Photosynthesis of the symbionts is the basis of the productive and species-rich coral reef ecosystem.
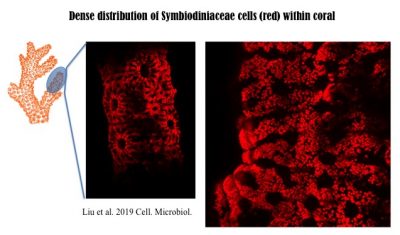
In corals, Symbiodiniaceae, typically of diverse genotypes (species) with one being dominant, are densely populated. The following 3D image was reconstructed from hundreds of Light Sheet Fluorescent Microscopic images. More technical information can be found in the recent paper published in Cellular Microbiology, https://doi.org/10.1111/cmi.13122, with video1, video2, video3, video4 available there.
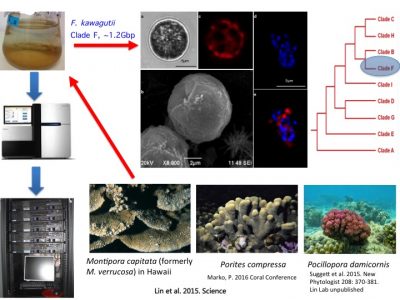
To understand molecular bases underpinning coral symbiosis, we sequenced the genome of Fugacium kawagutii, which was initially isolated from Montipora capitata in Hawaii, subsequently found in a couple of other corals (symbiotic relationship remains to be experimentally verified). F. kawagutii belongs to Clade F. The genome has undergone updates and is currently in V3, where assembly reaches chromosome level (one of them is 120Mbp long).
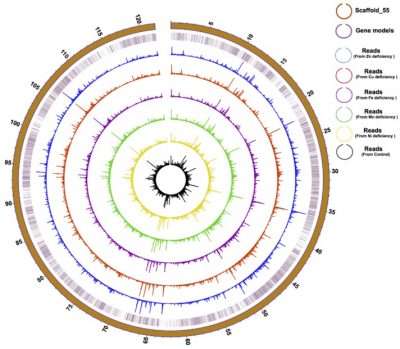
Physical distribution and expression profile of genes in the longest (121 Mbp) Scaffold in F. kawagutii. The circles from the outside to the inside mark scaffold length (in Mbp), gene location, and gene expression (bar height) under Zn-, Cu-, Fe-, Mn-, and Ni-deficient as well as normal conditions. See paper in Microorganisms, doi:10.3390/microorganisms8010102.
We examined how gene families differed from other non-symbiotic organisms: we found that 7663 gene families were gained and 338 were lost. Among the expanded gene families were those related to nutrient uptake and stress response. This makes sense because corals live in tropical habitats (warm with bright light and nutrient-poor environments, suspected to be stressful environments).
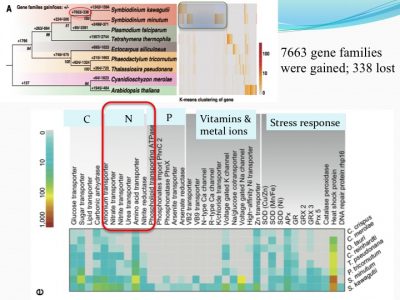
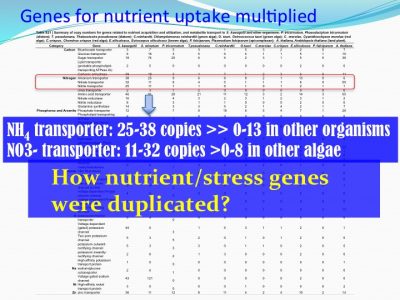
For instance, ammonium transporter and nitrate transporter genes are highly duplicated compared to other organisms.
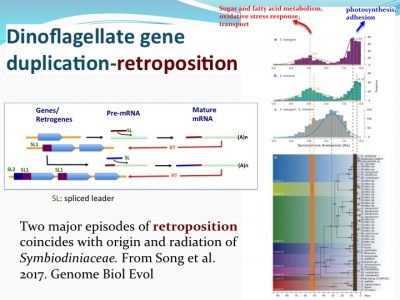
How nutrient/stress genes were duplicated? Dinoflagellates have an “unique” spliced leader (DinoSL) that provides a clue. The gene copies containing DinoSL at the 5′-end indicates they are duplicated through transposition. Genome wide survey of the transposed genes shows that during evolution, Symbiodiniaceae have experienced two major episodes of gene transposition, both coinciding with emergence and radiation of coral-Symbiodiniaceae symbiosis.
Many questions remain to be addressed regarding establishment, maintenance, and breakdown of dinoflagellate symbioses. Our lab uses a multi-faceted approach integrating different cutting-edge technologies to gain understanding of these processes.