UCONN Dino-Genomics Center:
Dinoflagellates are unicellular algae (heterotrophic protists for those without a chloroplast), forming a monophyletic group Alveolata with apicomplexans and ciliates. They are the second largest group of primary producers in the ocean (second to diatoms), indispensable for coral reef building and major contributors of harmful algal blooms. They possess two flagella and wear a cellulosic theca or are naked. Dinoflagellates possess a life cycle consisting of a vegetative stage reproducing by binary division, and cyst stages resulting from sexual fusion in response to unfavorable environmental conditions. They can be free-living in fresh or salty water, as plankton or sand dwellers and can be symbiotic or parasitic. About half of the 2000 extant dinoflagellates are heterotrophic ingesting other algae or dissolved organic matter (Schnepf and Elbrachter 1999), some of which can enslave ingested algal chloroplasts and perform ephemeral photosynthesis (e.g. Lewitus et al. 1999, Feinstein et al. 2002). These heterotrophic taxa are potentially important micrograzers in the microbial food web (Nakamura 1999). Some 60 taxa of dinoflagellates are known to form red tides, over 20 of which produce toxins that have profound impacts on fisheries industry, recreational values of coastal zones, and public health (Anderson 1994, 1996). For genomic database and analysis resources for dinoflagellates, please visit the SAGER website.
In evolutionary history, dinoflagellate genomes not only have undergone vertical evolution but have been impacted by rampant horizontal gene transfer. Dinoflagellates represent a photosynthetic organism with the most reduced plastid genome. The typical, peridinin-containing lineages have plastid genomes broken into single-gene minicircles that encode only 16 of the typically 130-200 plastid proteins (Koumandou et al. 2004). Transfer of the rest of the plastid genes to the nucleus has dramatically reconfigured the nuclear genome in dinoflagellates (Yoon et al. 2005). In addition, Rubisco has been replaced by that of proteobacterial origin (form II) likely through lateral gene transfer (Morse et al. 1995, Palmer 1996, Delwiche and Palmer 1996). Dinoflagellata is thought to lack histone proteins with few exceptions (e.g. Amoebophrya) (Rizzo 2003), but a histone H3-like protein (Okamoto and Hastings 2003) and a histone H2A.X (Hackett et al 2005) was reported recently in addition to findings of basic and acidic nuclear proteins (Hackett et al. 2005 and references therein, Zhang and Lin unpubl. data). Also, typical dinoflagellate cells divide with closed mitosis and extranuclear spindles, and chromosomes are permanently condensed. Other uncommon features for an eukaryote recognized so far include the rarity of mRNA splicing and deviation from the universal GT/AG rule (Palmer 1996), the extensive and novel mRNA editing in mitochondrial genes (Lin et al. 2002, 2008), and widespread spliced leader RNA trans-splicing (Zhang et al. 2007).
One most profound attribute of dinoflagellates is their huge genomes. In the past half century, the genome sizes of more than 30 dinoflagellates have been measured using various methods, giving a range of 3-278 pg DNA per genome or cell (e.g. Holm-Hansen 1969, Rizzo 1987, Veldhuis et al. 1997, LaJeunesse et al. 2005, etc), which is about 1-80 times that of the human haploid genome. Although smaller genomes may occur in some yet unrecognized dinoflagellates (Lin 2006), dinoflagellate genomes are still huge considering their relatively small cell size and being “simple” organisms, another case of C-value enigma (Gregory 2001). Equally striking is the wide range of genome size, which cannot be explained by conceivable difference in their apparent function or cell size. These peculiarities raise a question as to what portion of the dinoflagellate genome is protein coding and what function the remainder has. Early work on biochemical properties has shown that a large fraction of the nuclear DNA in the dinoflagellate Crypthecodinium cohnii and Prorocentrum cassubicum is composed of repeated and interspersed DNA sequences (Allen et al. 1975, Rizzo 1987, Hinnebusch et al. 1980, and Steele 1980). Hence, it has been suggested that the large fraction of the dinoflagellate genomes are nonfunctional (Anderson et al. 1992). We set out to gain a deeper level understanding of dinoflagellate genomics by sequencing the genome of the symbiotic dinoflagellate, Fugacium kawagutii.
Analysis of a Coral Symbionts Genome:
Coral reefs are the most productive and biodiverse marine ecosystems. This is because of the amazing symbiotic relationship between corals and the dinoflagellates from the family of Symbiodiniaceae. The two symbiotic partners benefit mutually. Photosynthesis of the symbionts is the basis of the productive and species-rich coral reef ecosystem.
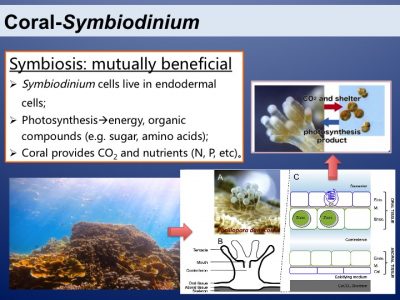
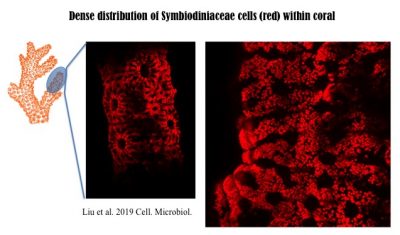
In corals, Symbiodiniaceae, typically of diverse genotypes (species) with one being dominant, are densely populated. The following 3D image was reconstructed from hundreds of Light Sheet Fluorescent Microscopic images. More technical information can be found in the recent paper published in Cellular Microbiology, https://doi.org/10.1111/cmi.13122, with video1, video2, video3, video4 available there.
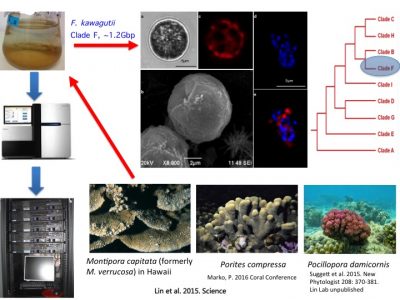
To understand molecular bases underpinning coral symbiosis, we sequenced the genome of Fugacium kawagutii, which was initially isolated from Montipora capitata in Hawaii, subsequently found in a couple of other corals (symbiotic relationship remains to be experimentally verified). F. kawagutii belongs to Clade F. The genome has undergone updates and is currently in V3, where assembly reaches chromosome level (one of them is 120Mbp long).
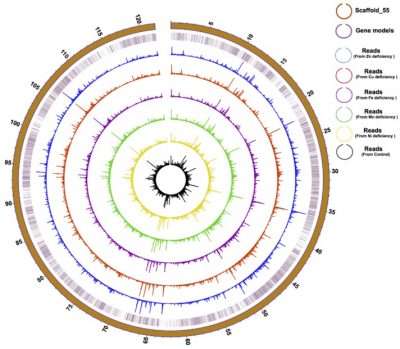
Physical distribution and expression profile of genes in the longest (121 Mbp) Scaffold in F. kawagutii. The circles from the outside to the inside mark scaffold length (in Mbp), gene location, and gene expression (bar height) under Zn-, Cu-, Fe-, Mn-, and Ni-deficient as well as normal conditions. See paper in Microorganisms, doi:10.3390/microorganisms8010102.
We examined how gene families differed from other non-symbiotic organisms: we found that 7663 gene families were gained and 338 were lost. Among the expanded gene families were those related to nutrient uptake and stress response. This makes sense because corals live in tropical habitats (warm with bright light and nutrient-poor environments, suspected to be stressful environments).
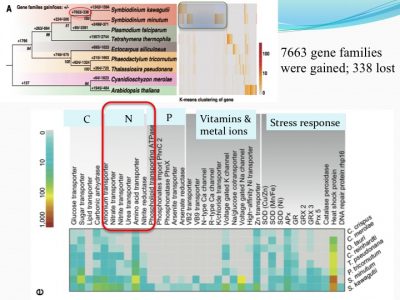
For instance, ammonium transporter and nitrate transporter genes are highly duplicated compared to other organisms.
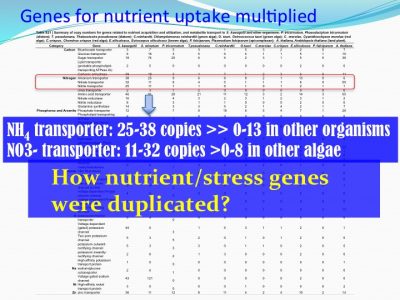
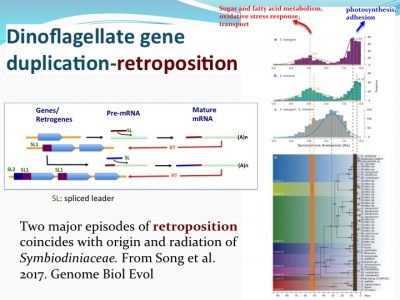
How nutrient/stress genes were duplicated? Dinoflagellates have an “unique” spliced leader (DinoSL) that provides a clue. The gene copies containing DinoSL at the 5′-end indicates they are duplicated through transposition. Genome wide survey of the transposed genes shows that during evolution, Symbiodiniaceae have experienced two major episodes of gene transposition, both coinciding with emergence and radiation of coral-Symbiodiniaceae symbiosis.
Many questions remain to be addressed regarding establishment, maintenance, and breakdown of dinoflagellate symbioses. Our lab uses a multi-faceted approach integrating different cutting-edge technologies to gain understanding of these processes.