These videos were taken three months after transformation using electroporation. The green fluorescence protein (GFP) was introduced using our DinoExpression vector (DinoIII-gfp). GFP is a protein that shows bright green fluorescence in the dark under UV and blue light wavelengths and is used in transformation techniques to serve as reporters, indicating visually that the introduced foreign DNA is being used. The GFP expressing Oxyhrris marina was swimming when taking this video.
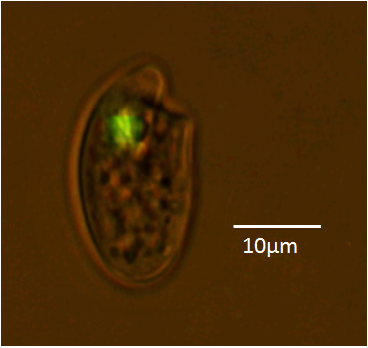
Here is a still image of a GFP expressing Oxyhrris marina
Dinoflagellates are a group of unicellular organisms (known as protists) living in aquatic environments. They have two flagella and many have theca (overlapping cellulose plates) as a cell wall. They are distributed widely, from polar to tropical waters. Their closest relatives are parasites such as Plasmodium (pathogens of malaria) and Perkinsus (pathogens of bivalves). Dinoflagellates include photosynthetic, heterotrophic, mixotrophic, and parasitic species. Photosynthetic species use sunlight and the green-house gas CO2 to make organic matter and latter becomes food for animals. Some of the photosynthetic species of dinoflagellates (in the genus of Symbiodinium) are essential endosymbionts of corals, and indispensable for coral reefs. Dinoflagellates also possess proteorhodopsin homologs suspected to supplement chloroplasts in harnessing solar energy. Dinoflagellates are particularly capable of utilizing dissolved organic phosphorus for phosphorus nutrient. Some species can emit light at night (bioluminescence). One manifestation of ecological success of dinoflagellates is their ability to grow to such a high abundance that changes the color of the water (commonly known as Red Tides), often inflicting devastating impacts on the coastal ecosystem, economy, and public health concerns (hence why they this phenomenon is named harmful algal blooms). Dinoflagellates are different in many ways from other organisms. They harbor many genes that we know nothing about their functions, and many “unusual” characteristics for which we have no clue what genes regulate. An effective tool of transformation will open a way to address this gap of knowledge.
It is a process to introduce a gene into a cell so that it will be expressed under the control of the host cell. If the expression of this “foreign” gene will cause changes in physiology or morphology that can be documented, the function of this gene can be revealed (forward genetics). By design, the expression of the introduced gene cassette can lead to editing of the host genome by disrupting a gene (commonly known as gene knockout or knockdown). If the knockout/knockdown of the gene will lead to a documentable change in physiology or morphology, the function of a an unknown gene can be unraveled (reverse genetics).
First, a proper vector is required to carry the gene. Typically a plasmid is used. The plasmid carries the target gene, regulatory elements to drive the expression of the gene, and genes that will facilitate selection of the transformed cells (selectable marker) from non-transformed cells (needed because transformation efficiency is usually low), and a reporter gene that allows researchers to determine that transformation is successful.
Second, the vector needs to be delivered into the target cell. There is a range of methods, mainly electroporation, biolistic, and bacterium-mediated (infection, conjugation, or otherwise co-incubation).
Third, selection of the transformed cells require a chemical agent. One kind of selection agent would kill the non-transformed cells, but not the the transformed cells because the vector carries a resistance gene. Another kind of selection is by a source of essential nutrient (e.g. N) that can be utilized only by the transformed cells because they express an enzyme (e.g. nitrate reductase) metabolizing the source compound.
Funded by the Gordon and Betty Moore Foundation-Marine Microbiology Initiative (MMI), the over goal of this endeavor is to develop a transformation tool that is suited to transform dinoflagellates. This include two major objectives:
- Screen strains of dinoflagellates for one or more tractable candidates for stable transformation and genomic manipulation.
- Develop a protocol that can be applied to the dinoflagellate strain(s) identified.

