Mitochondrial cytochrome b (cob) and cytochrome C oxidase subunit I (cox1) in dinoflagellates:
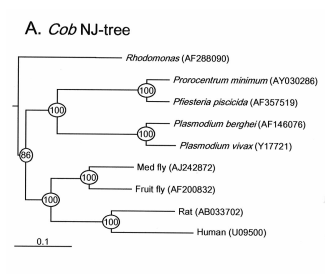
Mitochondrial cytochrome b gene has widely been used for phylogenetic analysis, population genetic structure analysis, and species identification for wide range of organisms. However, it has not been studied in dinoflagellates. Recently, we isolated this gene from the dinoflagellate Pfiesteria piscicida and examined its utility for phylogenetic analysis and species identification. Three copies of this gene were found, one of which appeared to be pseudogene. Neighbor-joining trees based on nt sequences exhibit more even pace of evolution across biological taxa than 18S rDNA. Based on the sequence, primers were designed one set of which was shown to be highly specific to P. piscicida. A Time-Step PCR protocol was established to quantify cell concentration of this species from both laboratory cultures and field-collected plankton samples. Appl. Environ. Microbiol. 68 (2002): 989-994.
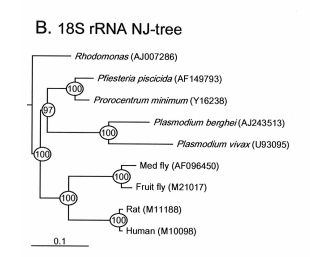
Pfiesteria Research:
Pfiesteria spp. are heterotrophic dinoflagellates that have been associated with fish kills in estuaries in North Carolina and Maryland Chesapeake Bay system. To date, two species have been identified, P. piscicida (meaning fish-killing) and P. shumwayae (named after Dr. Sandra Shumway). Although these organisms have been studied for a decade, still relatively little is known about this "mysterious" lineage. One of the focused research areas is to develop species-specifc tools for rapid identification and quantification of these organisms. Second important area is to study how environmental factors and genetic elements affect growth and regulate population of these organisms. Funded by ECOHAB, we have worked on these issues.
We developed a dual-gene PCR protocol for rapidly quantifying Pfiesteria piscicida with high sensitivity. We first isolated mitochondrial cytochrome b gene from P. piscicida, the first successful attempt working on this gene for dinoflagellates. We found that this gene is highly AT-rich, perhaps contributing to difficuties to clone in the past. Based on this gene sequence, species- specific primers were designed and used in PCR with another set of species-specific primers designed from the small subunit of ribosomal RNA gene (rDNA). With a Time-Step PCR technique, as few as 0.1 cell can be quantified.
Time Step PCR Showing Potential to Quantify P. piscidia Cell Concentration
It is important to have a second, independent, technique for quantifying P. piscicida cells in order to validate the PCR technique. In addition, there are circumstances where one technique may be preferable to the other. Therefore, we also developed an immunofluorescence technique (IF). Several antisera were produced in our laboratory and two of them proved to be very good (with titer of 3000 when staining on glass slides). IF is preferable when samples contain high level of PCR inhibitors and is easier to use. After tested against more than 20 species of algae, including P. shumwayae, these antisera appeared to be very species-specific.