Cyclins:
Cyclins by their names are known as proteins whose abundance oscillates in the cell division cycle. There are numerous cyclin proteins each of which is associated with a certain stage of the cell cycle. Cyclin B is one of them and is associated with mitosis (hence named mitotic cyclin). Cyclin B is synthesized in the S/G2 phases and its abundance peaks at G2/M transition. Cyclin B binds to p34cdc2 and induces activation of p34cdc2 by triggering the critical dephosphorylation (removing a phosphate group from some of the phosphorylated sites) of the kinase. The active p34cdc2-cyclin B complex functions as the mitosis promoting factor (MPF). Once mitosis is finished, cyclin B is dessociated from p34cdc2 and is degraded through a ubiquitin-dependent mechanism. Cyclin B is then synthesized again in the next cell cycle.
Cyclin B is conserved across wide range of taxa. A homolog in marine phytoplankton was recently detected and its encoded gene isolated (Lin et al. 1995; Lin and Zhang, in preparation)
p34cdc2:
p34cdc2 is a protein kinase which is activated when cells are committed to division. In eukaryotic cells, a cell undergoes G1 (gap or growth stage 1), S (DNA replication), G2 (gap or growth stage 2), and M (mitosis) phases leading to cell division. In typical mammalian cells, there are two checkpoints in the cell division cycle, the G1/S and G2/M transitions, respectively. When the cell is passing G1/S transition, it is considered "committed". Although p34 is shown widely to be present all the time, its activity oscillates through phosphorylation (getting a phosphate group at certain serine/tyrosine residues in the protein) and dephosphorylation (removing a phosphate group from some of the phosphorylated sites). The activated p34cdc2 bind with cyclin B and form a mitosis promoting factor (MPF) which trigers events of mitosis and cell division.
p34cdc2 is highly conserved across wide range of taxa. Its presence has been suspected earlier for freshwater alga like Chlamydomonas, this protein and its encoded gene was first detected and characterized for marine phytoplankton in our laboratory (Lin and Carpenter 1999).
PCNA:
Proliferating cell nuclear antigen (PCNA) is an accessory protein for DNA polymerase delta and is required for DNA replication. It is considered a "clamp" to enhance the processivity of the DNA polymerase. First found in human patients with autoimmune disease in 1985, this protein turned out to be ubiquitous and highly conserved. It has been detected in virus, archaea, and all eukaryotes. PCNA has been found to be associated with the S phase of the cell cycle. In eukaryotic cells, a cell undergoes G1 (gap or growth stage 1), S (DNA replication), G2 (gap or growth stage 2), and M (mitosis) phases leading to cell division.
In 1994, we first detected PCNA in marine phytoplankton. To date, PCNA and its encoding gene has been detected and characterized in a variety of phytoplankton taxa: chlorophyceae, bacillariophyceae, haptophyceae, cryptophyceae, pelagophyceae, and dinophyceae. We study phytoplankton PCNA because it can be used as a cell cycle marker for estimation of cell division rate in the environment. The underlying principle is that the completion of each round of the cell cycle results in growth of the population. Monitoring the progression of the cell cycle will allow one to calculate growth rate based on the time sequential values of the fraction of the cell population residing in this segment and the duration of the segment. Although PCNA marks the S phase, not quite near the M phase, our theoretical analysis shows that it can still provide a good approximation of in situ growth rate.
Cell Cycle Protein (CCP) Approach to In Situ Growth Rate Estimation:
Information on in situ growth rates (or cell division rates) is critical for understanding which of the processes, cell division (µ), import (I), grazing (G), cell death (M), or export (E), regulates the dynamics of phytoplankton populations.
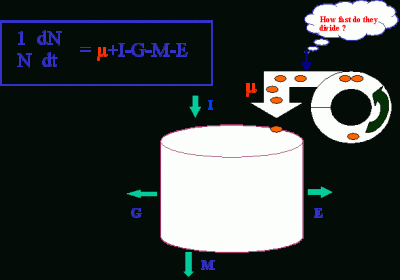
Practically, this information will be useful in assessing the contribution of a species (or a group of species) to carbon/nitrogen cycle, climate regulation, or harmful algal blooms.
In situ growth rate (µ) has been difficult to obtain due to lack of a feasible method. Over the past eight years, we have been developing a method named the cell cycle protein (CCP) method, which is based on the frequency of dividing cells technique (Weiler and Chisholm 1976, McDuff and Chisholm 1982) and the “terminal event” method (Carpenter and Chang 1988). All these share a common principle, that is the increase in population for the unicellular organism result from cell division. If the time sequential variation of the cell population that resides in a certain stage of the cell cycle can be determined along with the duration of this stage, µ can be estimated. The growth rate equation, developed for the “terminal event” method, is as follows,
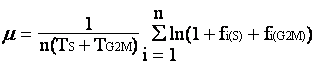
Basically, m is the growth rate (d-1), TS+TG2M is the duration of the terminal event, and fi(S)+fi(G2M) is the fraction of cells traveling through the S and G2-M phases in the ith sample. Advantages of this cell cycle approach are 1) no incubation of the organism is required; 2) growth rate estimates would not be affected by grazing, viral lysis, or physical transport of cells (assuming these processes are cell cycle independent).
There is a major improvement in the CCP method, which is the substitution of the presence of cell cycle proteins for the G2+M phase as the “terminal event” in the growth rate equation. Cell cycle proteins appear at specific stages in the cell division cycle and direct the cell to divide. Since they only appear in the cells that have reached a certain stage (e.g. G2+M phase of the cell cycle), their presence can be used to detect those cells. The presence of these proteins can be determined by using immunofluorescence on the whole cell with specific antibodies. The growth rate equation was modified (Lin et al. 1997) so that growth rate can be estimated regardless whether CCP-containing stages are terminal events. The equation now becomes:
eµ(Ts+TG2M) - eµTG2M - = 0
Where µ is the growth rate in d-1 to be resolved from this equation, TS and TG2M are the duration for the non-terminal event (represented by the S-phase) and the duration from this event to cell division (e.g. G2+M phase), respectively, and fS (bar) is the average fraction of a non-terminal event in the cell cycle over the diel sampling period. In the case that the cell cycle protein used represents a terminal event, i.e. TG2M = 0, the equation reduces to its earlier version (Eq. (1)). Alternatively, the expression of the CCP can be measured using reverse-transcription PCR, which is under investigation. The cell cycle proteins that have been explored include proliferating cell nuclear antigen (PCNA), p34cdc2, and cyclin B.